3.2 Get Spatial Data
ecodata::epu_sf
## Simple feature collection with 4 features and 3 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -77 ymin: 35.8327 xmax: -65.66667 ymax: 44.66667
## Geodetic CRS: NAD83
## EPU Shape_Leng Shape_Area geometry
## 0 GB 16.33086 6.162033 MULTIPOLYGON (((-66.5 42.16...
## 1 GOM 32.20684 7.545063 MULTIPOLYGON (((-69.26278 4...
## 2 SS 20.52712 3.350430 MULTIPOLYGON (((-67.54 44.6...
## 3 MAB 83.38407 15.695390 MULTIPOLYGON (((-75.97418 3...
epu_sf <- ecodata::epu_sf %>%
st_transform(4326)
class(epu_sf)
## [1] "sf" "data.frame"
# "sf" "data.frame"
g1 <- epu_sf$geometry[1]
#see in Environment pane, expand g1
plot(epu_sf)
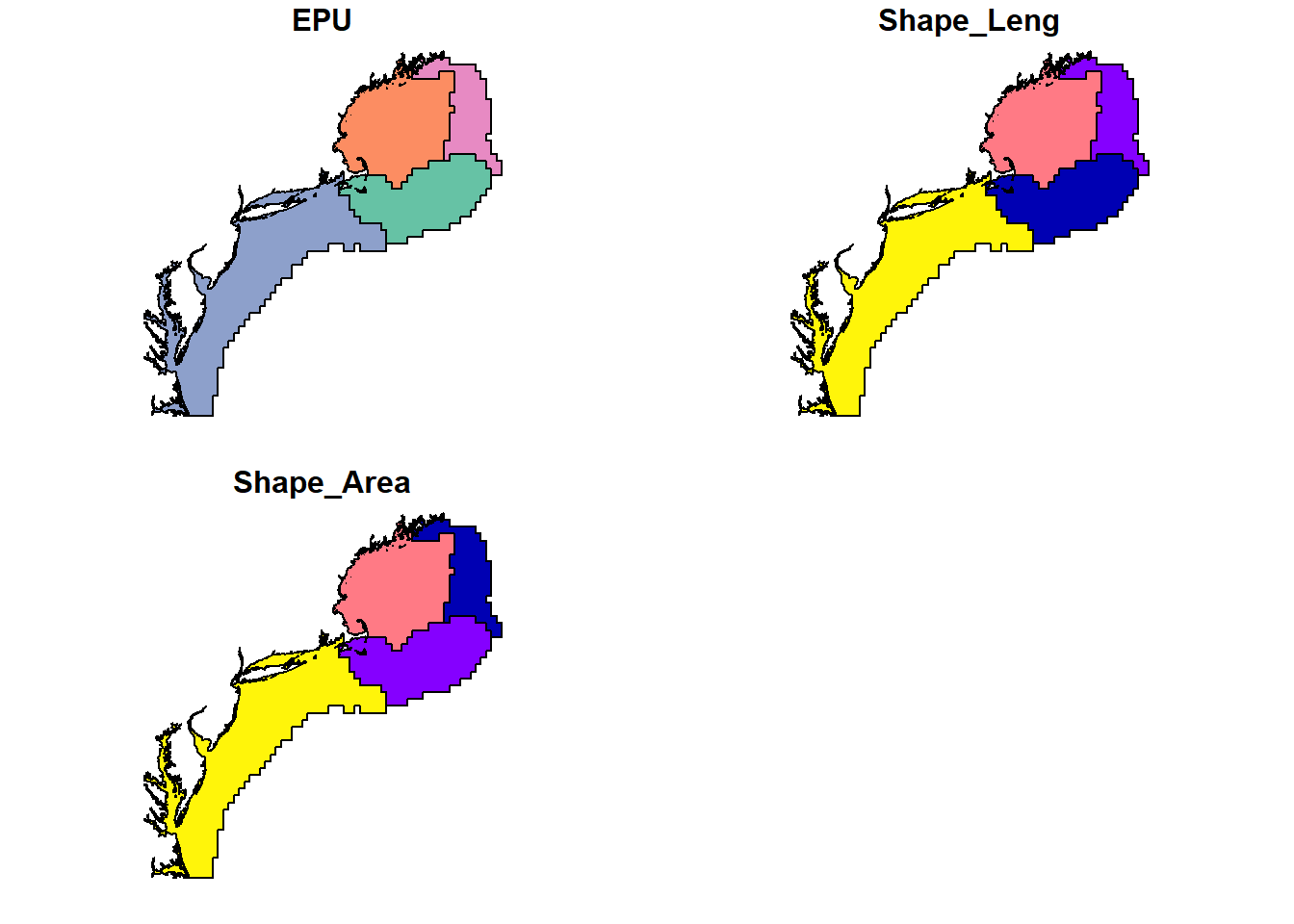
plot(epu_sf["EPU"])
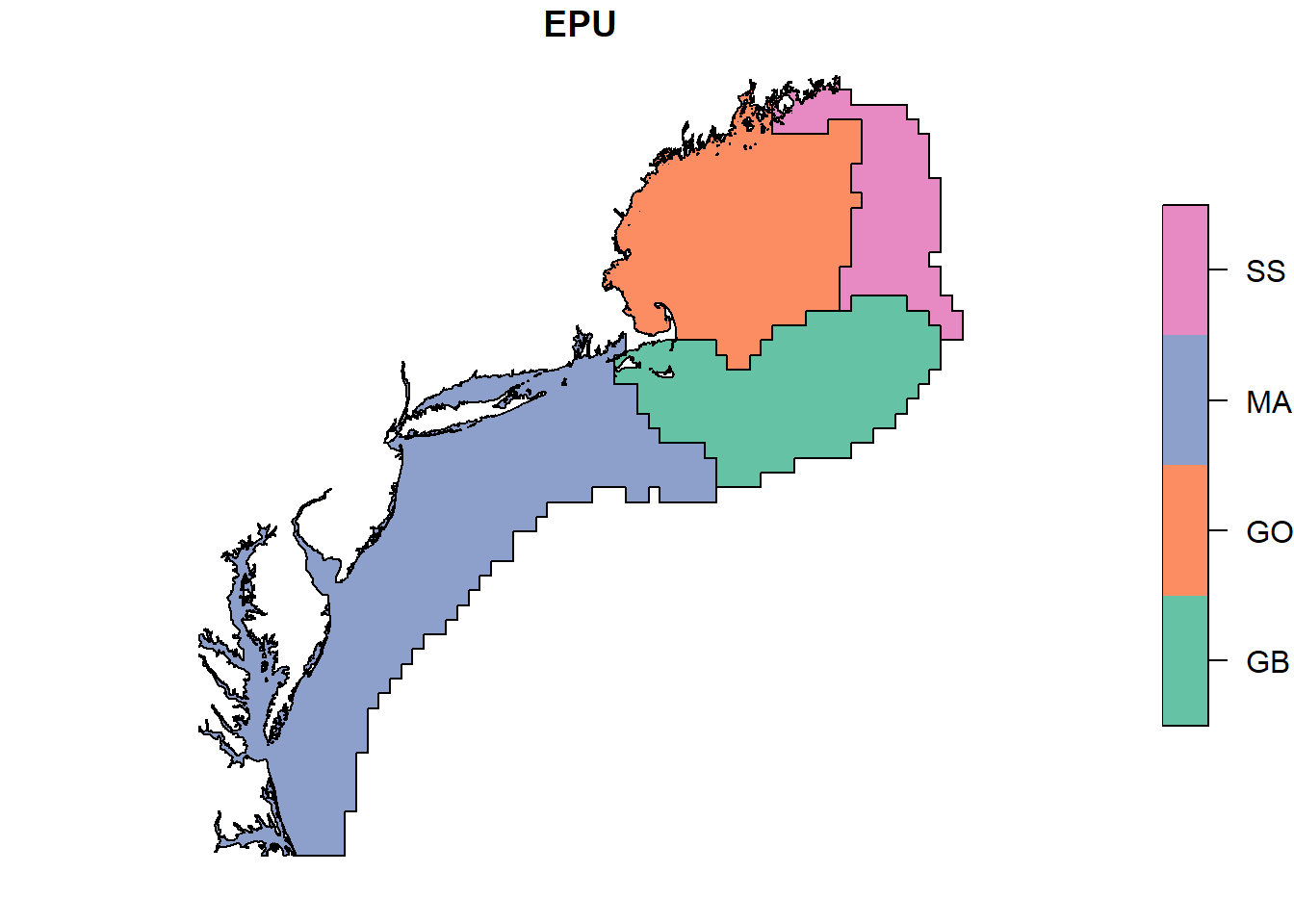
# to determine where this data falls on a map
shelf(mapview)
mapview(epu_sf)
# Mapping the data with shelf bathymetry data from the web
shelf(leaflet)
leaflet() %>%
#addTiles() %>%
addProviderTiles(providers$Esri.OceanBasemap) %>%
addPolygons(data=epu_sf)
using CoastWatch ERDDAP SST data from 2002-present
# install rerddap package for analysis
shelf(
here,
rerddap)
# get the data
sst_gd_rds <- here("data/sst_gd.rds")
epu_bb <- st_bbox(epu_sf)
epu_bb
## xmin ymin xmax ymax
## -77.00000 35.83270 -65.66667 44.66667
# looking at SST data
sst_info <- info('jplMURSST41mday')
sst_info
## <ERDDAP info> jplMURSST41mday
## Base URL: https://upwell.pfeg.noaa.gov/erddap/
## Dataset Type: griddap
## Dimensions (range):
## time: (2002-06-16T00:00:00Z, 2021-07-16T00:00:00Z)
## latitude: (-89.99, 89.99)
## longitude: (-179.99, 180.0)
## Variables:
## mask:
## nobs:
## sst:
## Units: degree_C
if (!file.exists(sst_gd_rds)){
sst_gd <- griddap(
sst_info,
fields="sst",
time=c("2020-06-16","2021-06-16"),
longitude=epu_bb[c("xmin","xmax")],
latitude=epu_bb[c("ymin","ymax")] )
saveRDS(sst_gd, file=sst_gd_rds)
}
sst_gd <- readRDS(sst_gd_rds)
sst_gd
## <ERDDAP griddap> jplMURSST41mday
## Path: [C:\Users\CORINN~1.PAT\AppData\Local\Cache\R\rerddap\6d0dbbf4f950f8dc4ffca5dd9b2cf661.nc]
## Last updated: [2021-08-04 13:18:50]
## File size: [52.2 mb]
## Dimensions (dims/vars): [3 X 1]
## Dim names: time, latitude, longitude
## Variable names: Sea Surface Temperature Monthly Mean
## data.frame (rows/columns): [13046670 X 4]
## # A tibble: 13,046,670 x 4
## time lat lon sst
## <chr> <dbl> <dbl> <dbl>
## 1 2020-06-16T00:00:00Z 35.8 -77 NA
## 2 2020-06-16T00:00:00Z 35.8 -77.0 NA
## 3 2020-06-16T00:00:00Z 35.8 -77.0 NA
## 4 2020-06-16T00:00:00Z 35.8 -77.0 NA
## 5 2020-06-16T00:00:00Z 35.8 -77.0 NA
## 6 2020-06-16T00:00:00Z 35.8 -76.9 NA
## 7 2020-06-16T00:00:00Z 35.8 -76.9 NA
## 8 2020-06-16T00:00:00Z 35.8 -76.9 NA
## 9 2020-06-16T00:00:00Z 35.8 -76.9 NA
## 10 2020-06-16T00:00:00Z 35.8 -76.9 NA
## # ... with 13,046,660 more rows
names(sst_gd)
## [1] "summary" "data"
# mapping most recent SST values with a gradient
shelf(
dplyr,
ggplot2,
mapdata)
#coastline
coast <- map_data(
"worldHires",
xlim=epu_bb[c("xmin","xmax")],
ylim=epu_bb[c("ymin","ymax")],
lforce="e")
sst_df_last <- sst_gd$data %>%
filter(time == max(time))
# summary(sst_df_last)
ggplot(
data=sst_df_last,
aes(x=lon, y=lat,fill=sst)) +
geom_polygon(
data=coast,
aes(x=long,y=lat,group=group), fill="grey80" ) +
geom_tile() +
scale_fill_gradientn(
colors=rerddap::colors$temperature,na.value=NA ) +
theme_bw() +
ylab("Latitude") +
xlab("Longitude") +
ggtitle("Latest SST")
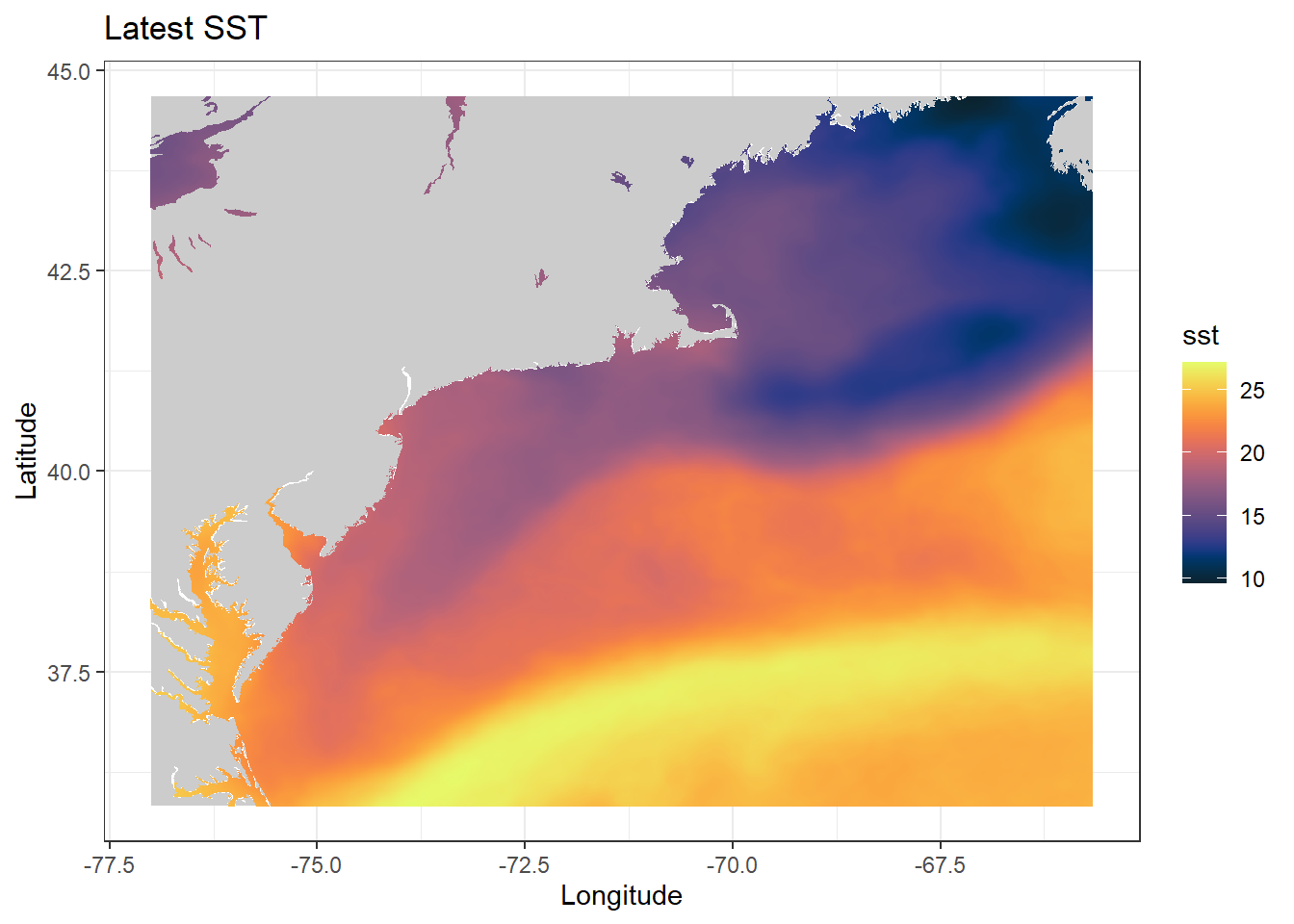
shelf(
purrr, raster, sp, tidyr)
select <- dplyr::select
#generate tibble of SST data
sst_tbl <- tibble(sst_gd$data) %>%
mutate(
#round b/c of uneven intervals
# unique(sst_gd$data$Lon) %>% sort() %>% diff() %>% table()
lon=round(lon,2),
lat=round(lat,2),
date=as.Date(time,"%Y-%m-%dT00:00:00Z") ) %>%
select(-time) %>%
filter(!is.na(sst)) #13M to 8.8M rows
#convert to monthly data
sst_tbl_mo <- sst_tbl %>%
nest(data=c(lat,lon,sst)) %>%
mutate(
raster=purrr::map(data,function(x) {
#browser()
sp::coordinates(x) <- ~lon + lat
sp::gridded(x) <- T
raster::raster(x)
}) )
#stack the monthly raster data
sst_stk <- raster::stack(sst_tbl_mo$raster)
names(sst_stk) <- strftime(sst_tbl_mo$date, "sst_%Y.%m")
raster::crs(sst_stk) <- 4326
shelf(stringr)
# extract the mean and standard deviation of SST across time
epu_sst_avg <- raster::extract(sst_stk, epu_sf, fun=mean, na.rm=T)
epu_sst_sd <- raster::extract(sst_stk, epu_sf,fun=sd, na.rm=T)
# transform those values to a tibble
epu_sst_tbl <- rbind(
epu_sst_avg %>%
as_tibble() %>%
cbind(
EPU = epu_sf$EPU,
stat="mean" ) %>%
pivot_longer(-c(EPU,stat)),
epu_sst_sd %>%
as_tibble() %>%
cbind(
EPU=epu_sf$EPU,
stat="sd" ) %>%
pivot_longer(-c(EPU, stat))) %>%
mutate(
EPU=as.character(EPU),
date=as.double(str_replace(name,"sst_","")) ) %>%
select(-name) %>%
pivot_wider(
names_from = EPU,
values_from = value )
# graphing the tibble of mean SST over time
shelf(dygraphs)
epu_sst_tbl %>%
filter(stat == "mean") %>%
select(-stat) %>%
dygraph()


